Outline of the Research
Organisms have adapted over a long period of time by developing mechanisms to eliminate invasive RNAs or suppress the expression of RNAs that perturb their own systems, and sometimes by co-opting them. However, with the frequent emerging viral infections in recent years and the practical application of artificial mRNAs, the opportunities to accept RNAs whose medium- to long-term effects are unclear are rapidly increasing. To address these pressing issues, we define perturbing RNAs (perRNAs) as a group of RNAs that disrupt living systems, whether exogenous or endogenous, and aim to characterize them by bringing together researchers from different fields to comprehensively understand the regulatory mechanisms of perRNAs and their disruption in disease and adaptation to environmental changes. This attempt to reconsider RNA from the viewpoint of negative functions will bring about a paradigm shift from preconceived notions about RNA. The results of this project will be widely disseminated as a “perRNA database” to share the novel concept of perRNA with researchers in Japan and overseas. This will make it possible to elucidate perRNAs associated with a wide range of species, diseases, and environmental changes, and to predict their physiological effects with a high degree of accuracy. As a result, physiological phenomena, causes of diseases, and adaptation mechanisms to environmental changes will be clarified, leading to the development of new disease treatments and RNA medicines with fewer adverse reactions.
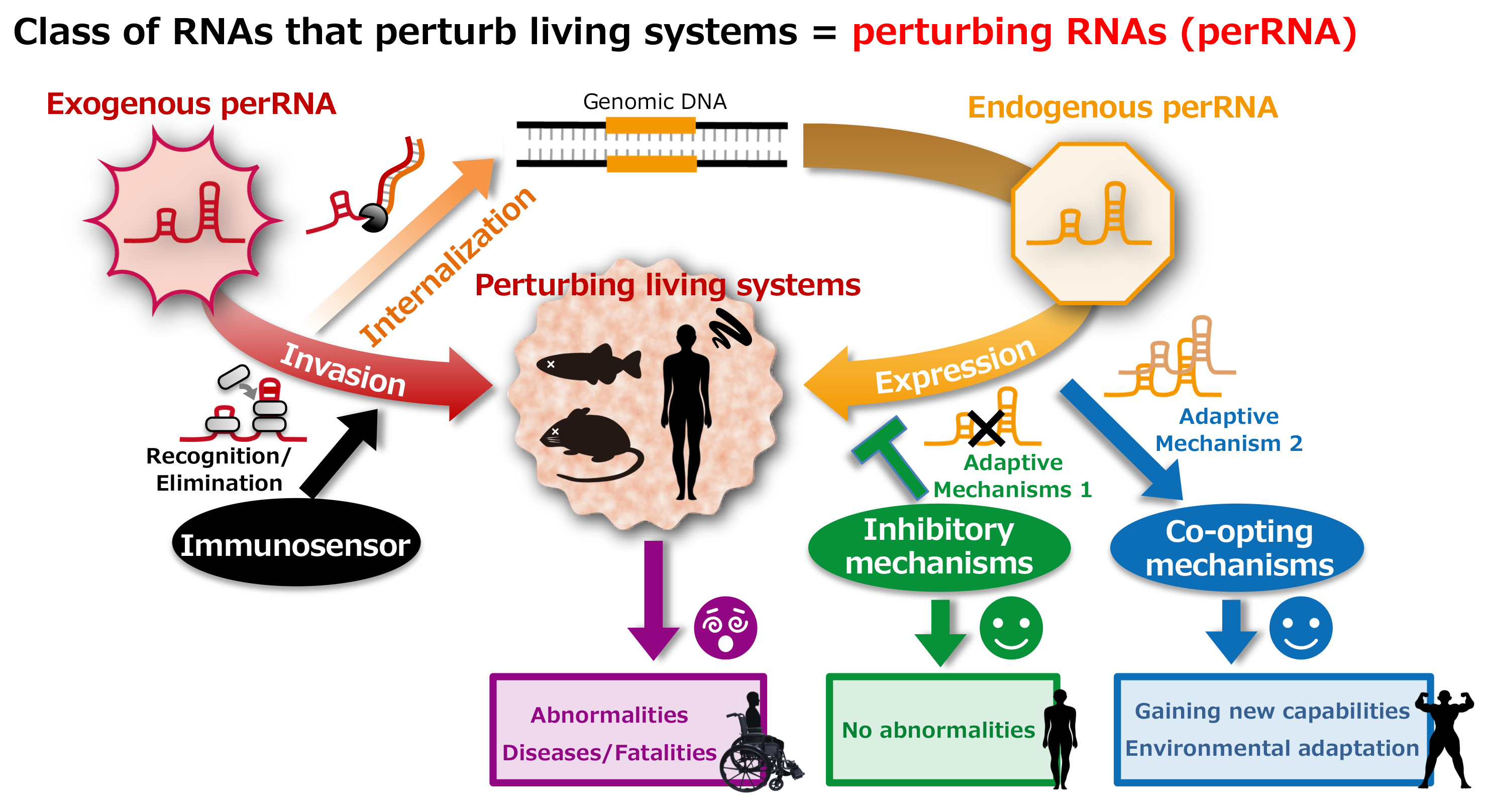
Background
In recent years, emerging RNA viral infections such as COVID-19 have increased in the human population, and these viral RNAs perturb the host’s biological systems, sometimes causing cellular and individual death.
In addition, artificial RNAs, as represented by mRNA vaccines, are expected to be used with increasing frequency, but the medium- to long-term effects of their introduction are unknown. On the other hand, the majority of biological genomes are composed of sequences derived from foreign RNAs. Retrotransposons, a prime example, have given rise to the expression of endogenous RNAs, are being elucidated to have various functions other than transposition. Furthermore, the risk of double-stranded RNA formation between inactive repeat sequences has increased, and it has become essential to distinguish them from exogenous double-stranded RNAs such as viruses. It is assumed that the organism has multiple layers of inhibitory mechanisms as adaptive measures against such endogenous perRNAs. In addition, it is becoming aware of some cases of co-opting RNAs derived from retrotransposon, but much of these mechanisms remain unresolved.
The goal of this research field is to understand the adaptive mechanisms that enable coexistence with perRNAs. This ambitious project aims to comprehensively reconsider RNAs from the viewpoint of negative functions, bringing about a paradigm shift from preconceived notions about RNA.
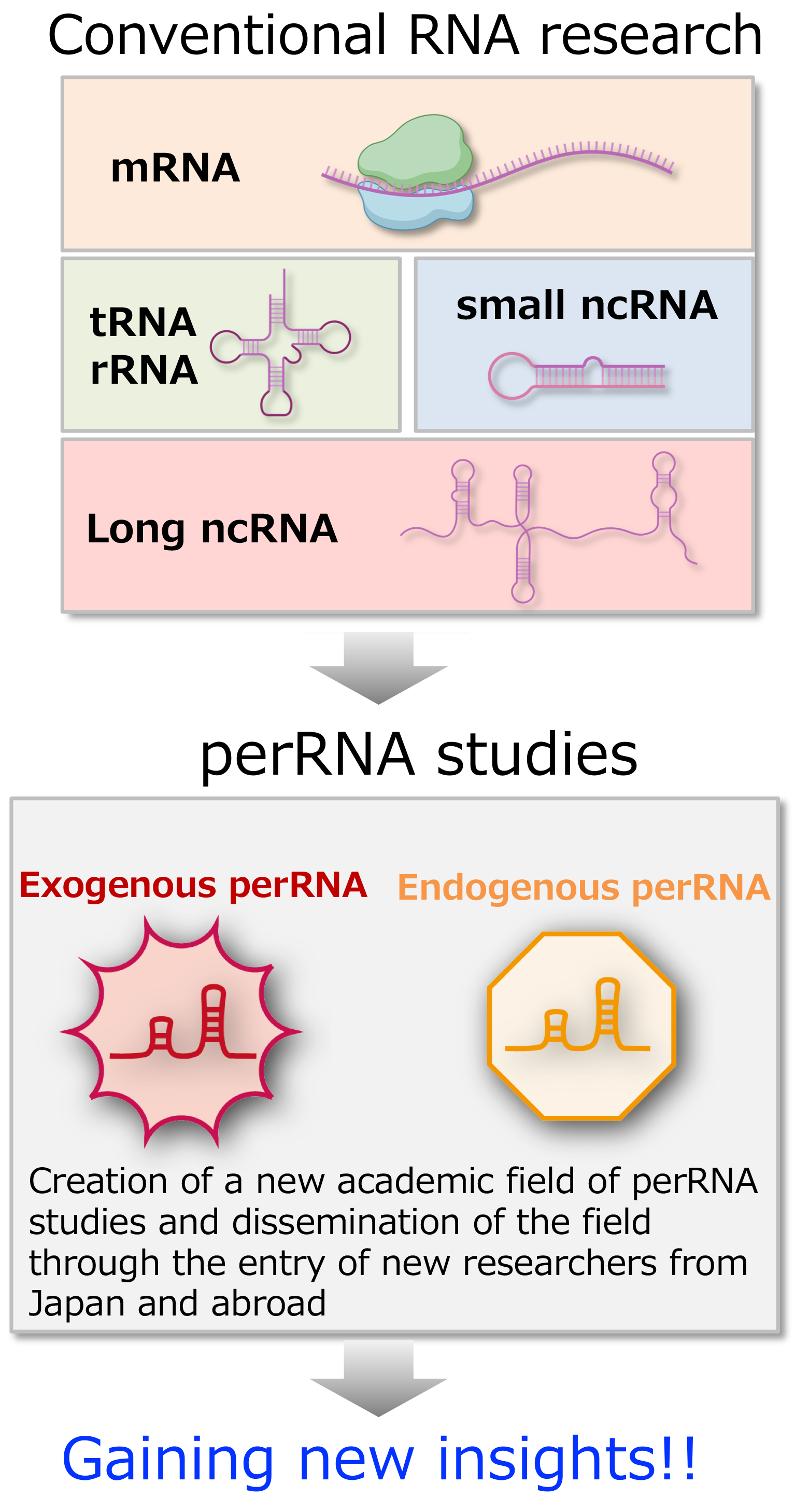
Research contents
Project A01: Identification and characterization of perRNAs
From public databases and transcriptome data obtained by each group’s research, perRNA candidates with specific functions will be extracted and characterized in terms of sequence, structure, and dynamics.
Project A02: Elucidation of molecular mechanisms that manipulate perRNAs
To understand perRNA adaptation mechanisms, we will elucidate the mechanisms that manipulate perRNA functions. Essentially, perRNAs that disturb cellular and individual homeostasis are suppressed so that their functions are not exerted. To this end, it is essential to understand the recognition mechanisms that sense perRNAs, induce their degradation, and suppress their expression and function. We will also the mechanisms underlying co-opted perRNAs.
Ripple effects
The outcomes of each research group will be registered in the “perRNA database”, which will enable the prediction of physiological effects of perRNAs in a wide range of species, diseases, and environmental changes with assistance of AI. The accumulation of such knowledge will provide an opportunity to reconsider RNAs from the viewpoint of negative functions, bringing about a paradigm shift from preconceived notions about RNA. As a result, it is expected to be applied to the development of new disease therapies and RNA medicines with fewer adverse reactions.
